Metabolism, epigenetics and breast cancer
Research Project
The Havas lab investigates metabolic reprogramming during the initiation, progression and response of breast cancer using in vivo animal model systems and ex vivo organotypic cultures. We aim to understand how both genetic context and the local environment, shape metabolic reprogramming in breast cancer. Specifically, we are interested in understanding the extent of metabolic plasticity throughout the evolution of breast cancer, from disease onset up to and including therapeutic response. Through more thorough understanding of this key hallmark of breast cancer we aim to design strategies for more effective patient treatment.
Modeling minimal residual disease
Breast cancer is the most frequently diagnosed cancer among women, with more than half-a-million women succumbing to breast cancer annually world-wide. Mortality is largely due to tumor recurrence, which is attributed to therapy resistant tumor cells. The identification and characterization of the surviving tumor cells is a major challenge in the field. We use a combination of mouse models and organoid cultures to isolate cells which survive therapeutic treatment (Havas et al., submitted). This approach has allowed us to characterize residual breast cancer using multi-tiered systems biology approaches. Our goal is to utilize these platforms to identify more effective therapeutic options for breast cancer patients
Addicted to metabolism
Therapeutic efficacy is largely hampered by intra-tumoral heterogeneity.Yet, despite transcriptional and mutational heterogeneity cancer cells derived from diverse tumor types often have similar metabolic abnormalities. The specificity of these metabolic alterations to cancer cells makes them attractive therapeutic targets. Yet, there remains a large gap in the understanding of how both genetic and local environmental factors shape tumor metabolism. Using a combination of computational and wet lab approaches we have recently uncovered lipid metabolic dependencies in breast cancer (Havas et al.,submitted). We are now moving forward utilizing the power of genome-editing approaches to identify key regulators of these pathways in breast cancer, so that we may better treat malignant disease.
Defining (epi)genetic drivers of breast cancer
It has been reported that several chromatin-modifying enzymes require intermediates of cellular metabolism for their function. This is most clearly illustrated at the level of histone post-translational modifications, which are regulated by enzymes that are either regulated by, or catalyze the transfer of, metabolic intermediates to histones. Post-translational chemical modification of chromatin sensitizes the genome to changing environmental and physiological conditions, and has been shown to play an important role in cancer progression. Yet, the role of cancer specific metabolic intermediates in shaping epigenetic transformation has only recently begun to be addressed. Using inducible model systems we are exploring the cross talk between metabolism and the epigenetic transformation during the establishment, development, and treatment of breast cancer.
Photogallery
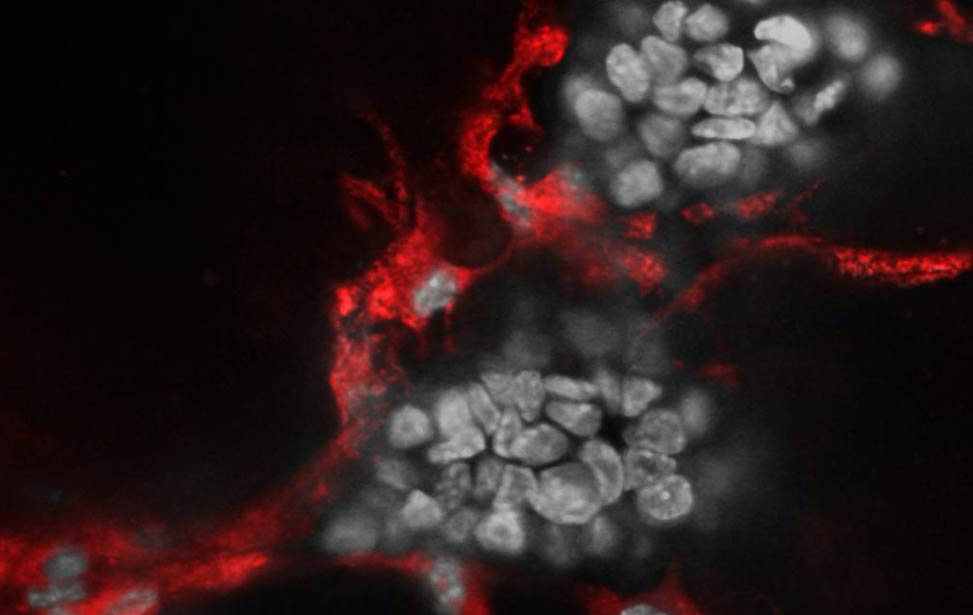 Co-culture of Her2/neu expressing organoid(grey) and primary endothelial cells(red)
Co-culture of Her2/neu expressing organoid(grey) and primary endothelial cells(red)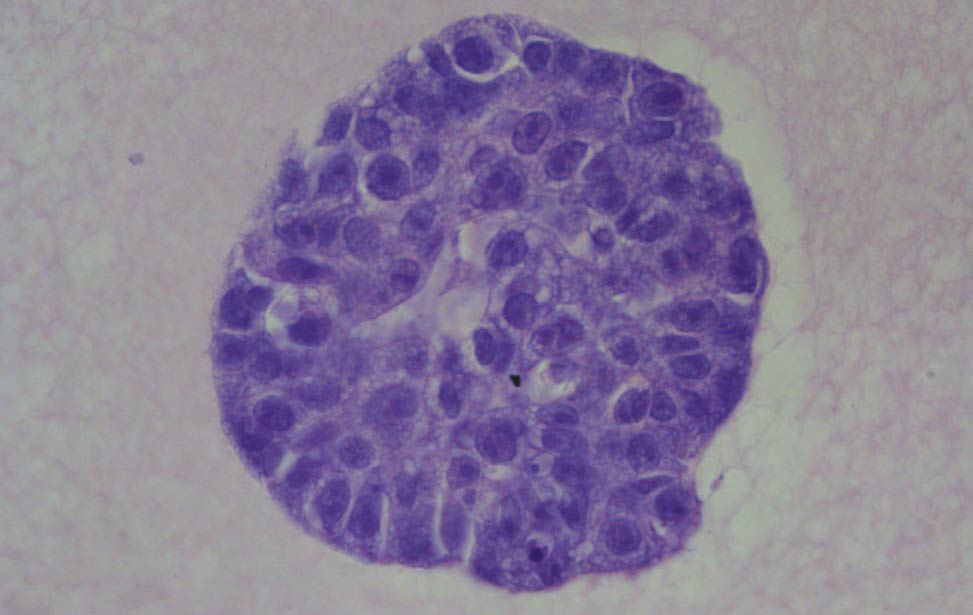 Hematoxylin and eosin staining of organoid expressing Her2/neu
Hematoxylin and eosin staining of organoid expressing Her2/neu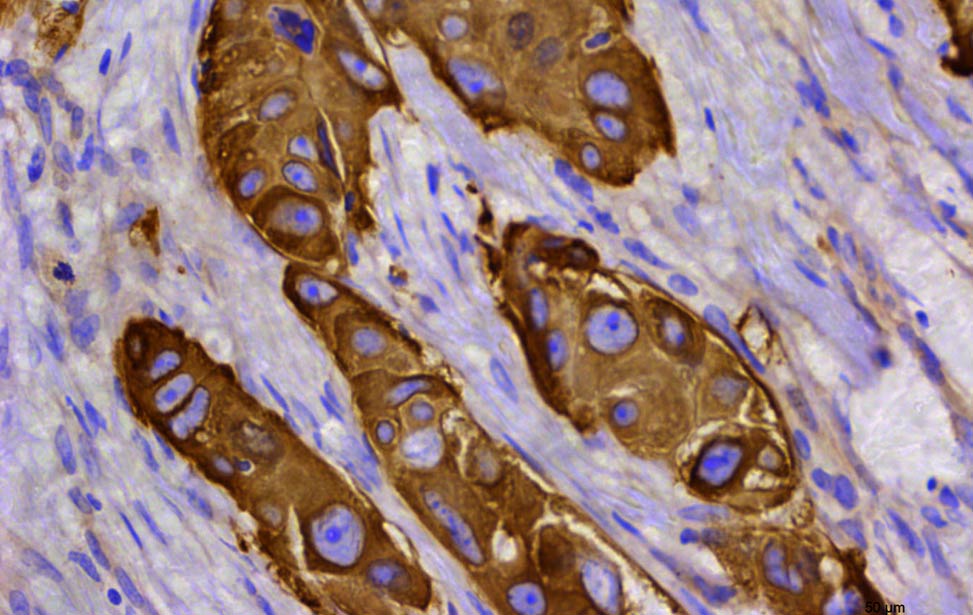 FASN expression in human breast tumor
FASN expression in human breast tumor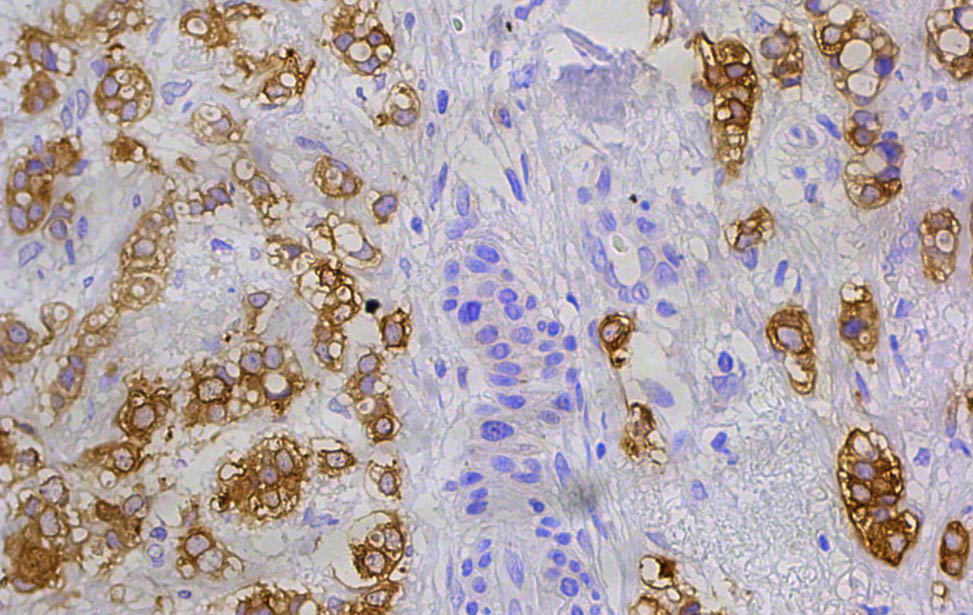 FASN expression in human breast tumor
FASN expression in human breast tumor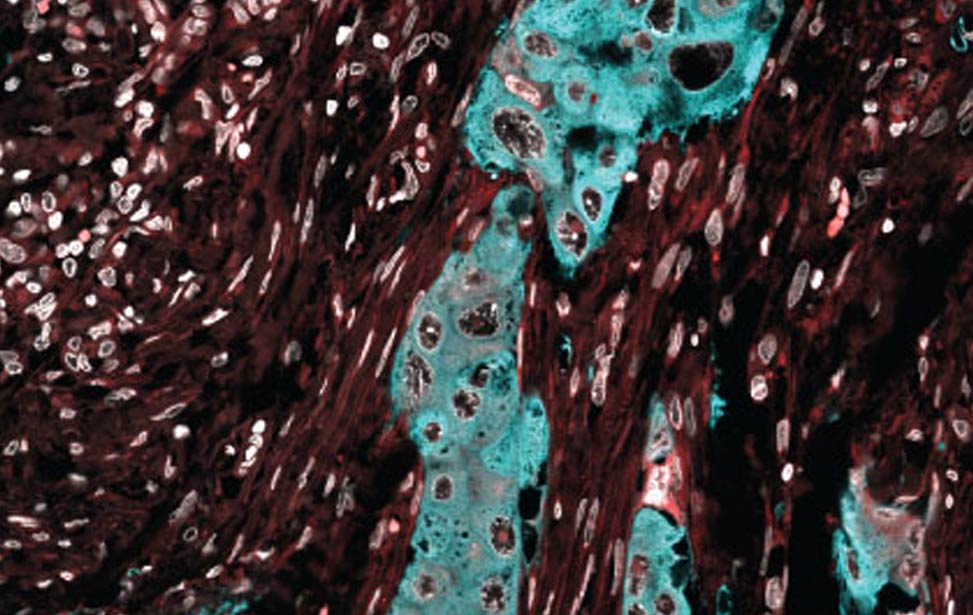 FASN expression (cyan) in human breast tumor
FASN expression (cyan) in human breast tumor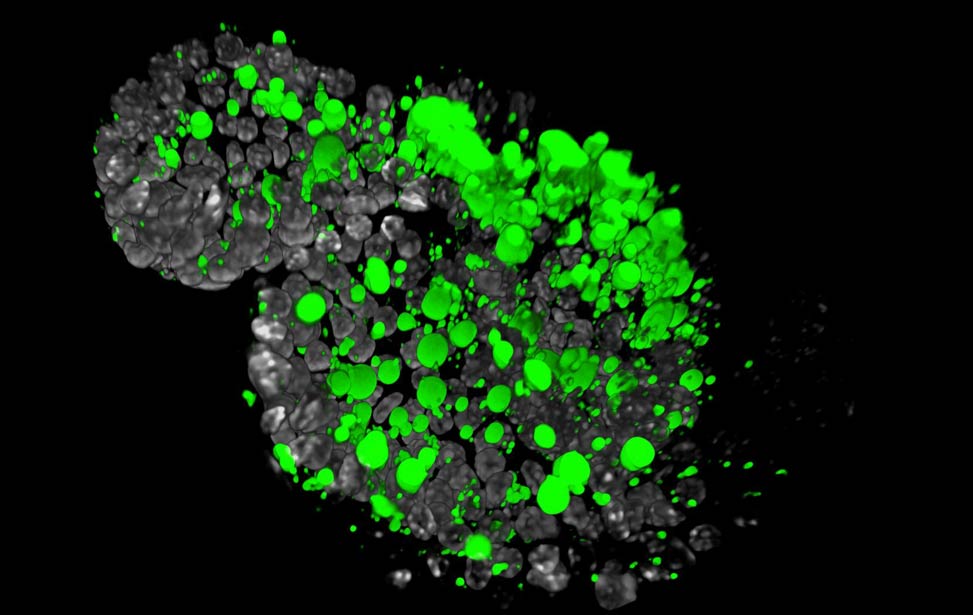 Lipid droplets (green) in Her2/neu expressing
organoid
Lipid droplets (green) in Her2/neu expressing
organoid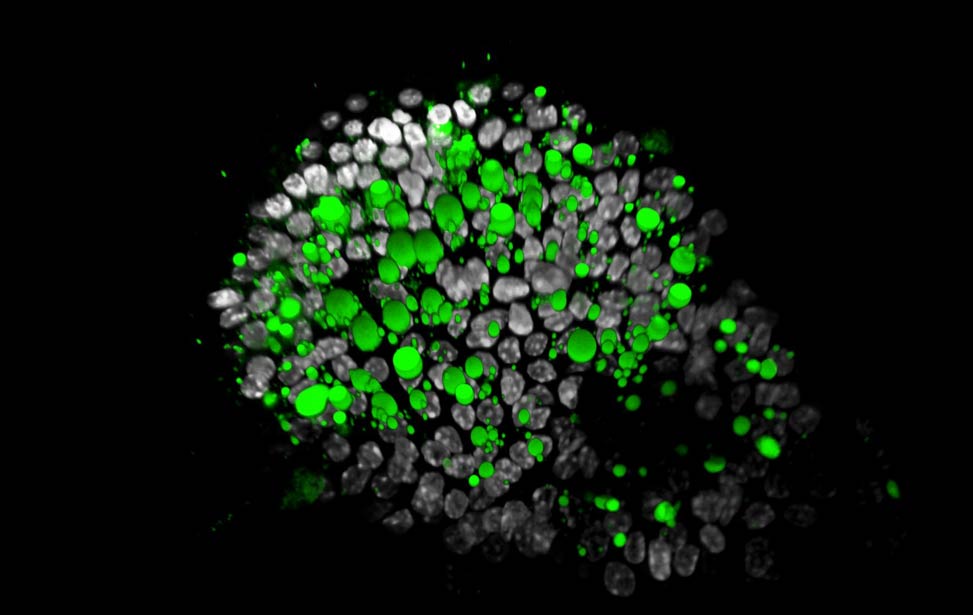 Lipid droplets (green) in Her2/neu expressing
organoid
Lipid droplets (green) in Her2/neu expressing
organoid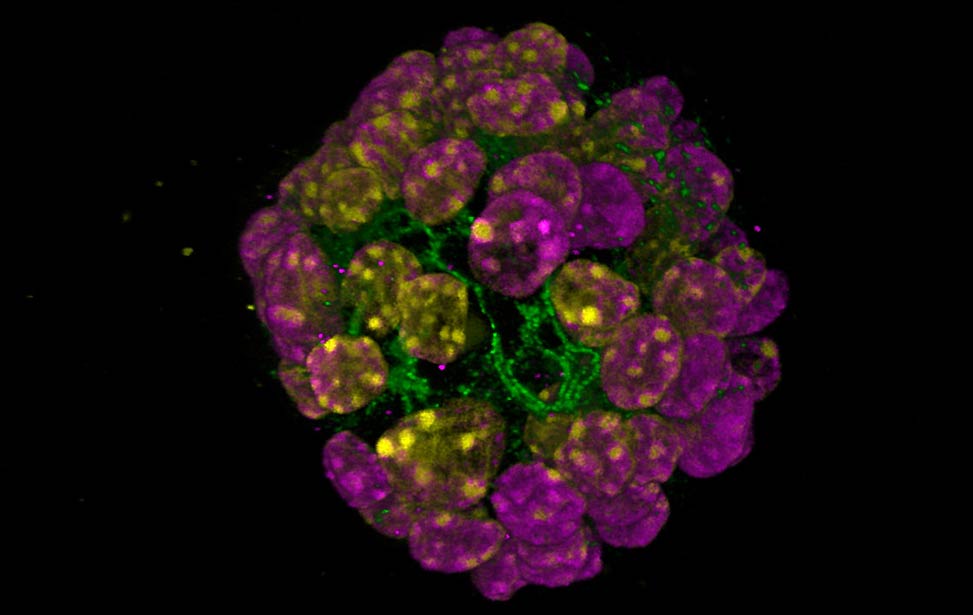 H3K9me3(magenta) and H3K4me3(yellow) staining of organoid
expressing Her2/neu
H3K9me3(magenta) and H3K4me3(yellow) staining of organoid
expressing Her2/neu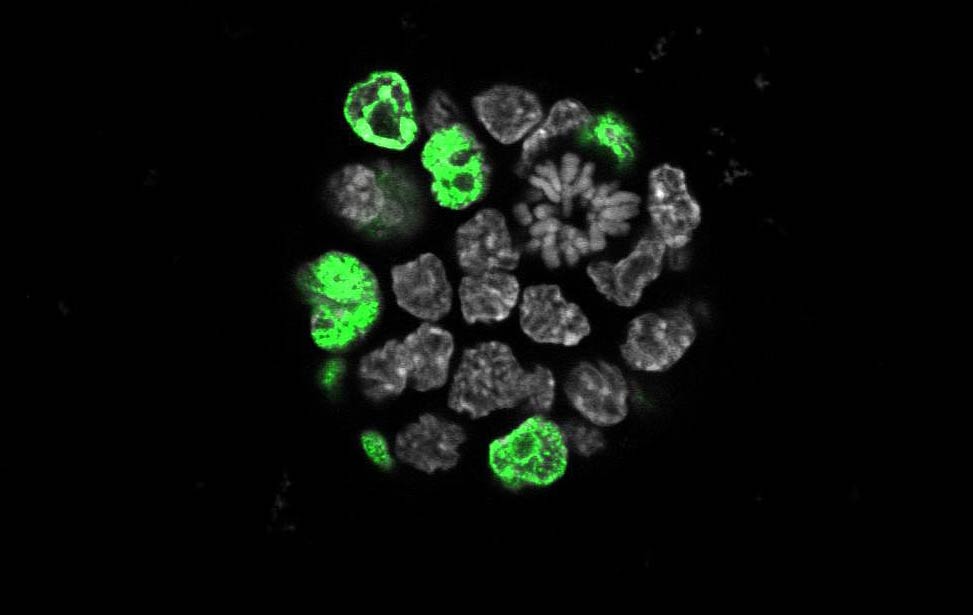 EdU staining for proliferation in an organoid expressing Her2/neu
EdU staining for proliferation in an organoid expressing Her2/neu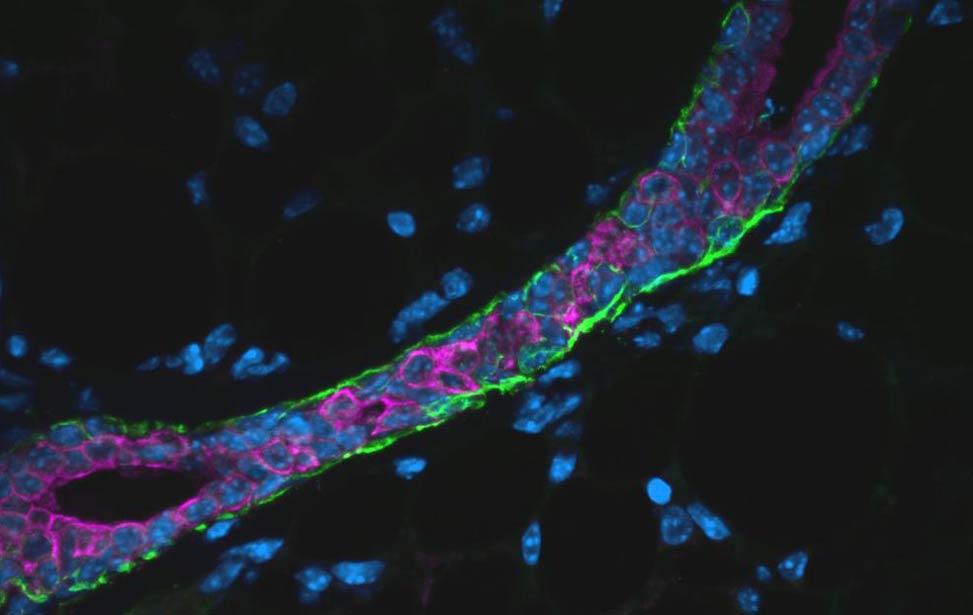 Immunofluoresence staining of section of mouse mammary gland
[SMA(green) and TROMA(magenta)]
Immunofluoresence staining of section of mouse mammary gland
[SMA(green) and TROMA(magenta)]