DNA metabolism
The role of DNA damage repair and DNA damage response genes in vertebrate organisms
Cells respond to DNA damage and replication stress, by activating a multi-faceted response called DNA damage response. In contrast to lower eukaryotes many genes involved in DNA repair and in DNA damage response are lethal when inactivated in mammalian cells even in the absence of apparent DNA damage. This indicates that these genes fulfil essential tasks for cell survival in complex organisms. In addition, many genes controlling DNA metabolism such as BRCA1/2, the Fanconi Anemia proteins and p53 are only present in metazoan organisms implying a higher intrinsic complexity of DNA metabolism.
Defects in DNA repair and DNA damage response can result in genomic instability, a major feature of cancer cells. Several cancer-prone syndromes reflect defects in the specific genes of the DNA damage response. Several players of the DNA damage response and DNA repair processes are known. However, their biochemical function is poorly understood. Furthermore, the role of DNA repair and DNA damage response in stem cells is largely unknown.
In the past, we identified novel ATM and ATR DNA damage checkpoints that prevent initiation of DNA replication following DNA damage (Costanzo et al Molecular Cell 2000 and 2003).
We studied DNA repair pathways and identified novel functions for the Mre11 complex that account for its essential role in vertebrates (Costanzo et al Molecular Cell 2001).
More recently, we have dissected the molecular mechanisms underlying RAD51 and Mre11 function during DNA replication using new methodologies that we developed based on Electron Microscopy (EM) to visualize replication intermediates derived from egg extract. We demonstrated that Mre11 promotes degradation of nascent strands at stalled forks and that RAD51 protects replication forks from this processing (Hashimoto et al Nature Structural and Molecular Biology 2010).
We have also shown that ATM regulates the pentose phosphate pathway promoting the formation of antioxidant molecules such as NADPH and nucleotide precursors required for DNA repair (Cosentino et al EMBOJ 2006).
We have isolated genes present exclusively in vertebrate organisms such as GEMC1 and Cep63 that regulate essential cell cycle transitions such as initiation of DNA replication and mitosis spindle assembly under perturbed and unchallenged conditions (Smith et al Nature Cell Biology 2008; Balestrini et al Nature Cell Biology 2009).
We have shown that centromeric repetitive DNA assembles functional centromeric chromatin, which forms DNA loops and suppresses the activation of ATR (Aze et al Nature Cell Biology 2016).
Finally, we have demonstrated that BRCA2-RAD51 coordinate the function of DNA polymerases at difficult to replicate DNA regions, preventing the accumulation of ssDNA gaps at stalled forks. Such gaps can be transformed in reversed forks by the action of SMARCAL1 translocase. Reversed forks trigger extensive nascent DNA degradation in the absence of the protective function of RAD51, which directly prevent DNA degradation operated by DNA nucleases (Kolinjivadi et al Molecular Cell 2017).
Current research projects
The ongoing projects of my group aim at understanding the molecular roles played by vertebrate DNA metabolism genes in various aspects of cell cycle, DNA replication and DNA repair in unchallenged and stressful conditions. In particular, we are studying:
- The ATR dependent response to replication stress at molecular, cellular and organism level in vertebrates
- The molecular role of DNA repair and replication proteins BRCA1/2, RAD51, PARP and DNA polymerases at normal and stalled replication forks
- The ATM dependent signal transduction pathway and its links with cellular metabolism in normal cells and in Ataxia Telangiectasia
- The role of DNA repair and DNA damage response proteins in vertebrate stem cells, embryonic development and cancer
To this end, we are applying a multidisciplinary approach. In particular, we are taking advantage of cell-free extracts, mass-spectrometry analysis of protein-protein interaction networks, antibody based techniques, mammalian cell manipulation, mouse genetics and cells from patients affected by Ataxia-Telangiectasia as tools and model systems.
The Xenopus laevis model facilitates the biochemical study of large essential proteins that can be easily isolated and characterised. An important tool available with this system is the possibility of depleting native proteins and protein complexes using specific antibodies. This approach allows the identification of the precise biochemical steps in which these proteins operate allowing the study of essential vertebrate genes that would normally compromise viability when inactivated in other cellular systems. This system offers unique opportunities not available with other known experimental models to define the molecular role of essential DNA metabolism proteins.
More recently we have started to investigate the role of DNA repair proteins in mouse embryos applying the same concepts that have guided our investigation in other model systems. This approach is complemented by the use of mass spectrometry analysis and advance imaging approaches, including electron and atomic force microscopy (AFM) based analysis of DNA metabolism intermediates. These studies are aimed at understanding biological and biochemical functions of genes involved in essential DNA repair processes that are dysfunctional in cancer and aging cells.
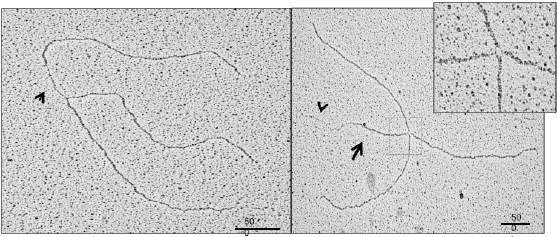
Figure 1. EM image of replication fork isolated from cells defective for BRCA2 showing a ssDNA gap (arrow) at fork junction (left). A typical reversed fork induced by replicaiton fork stalling (right) (Kolinjivadi et al, Molecular Cell 2017)
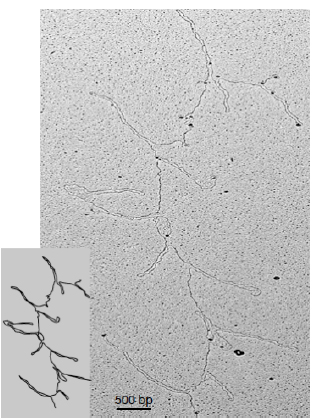
Figure 2. EM image of DNA loops formed by centromeric chromatin assembled with human repetitive centromeric DNA (Aze et al. Nature Cell Biology 2016)
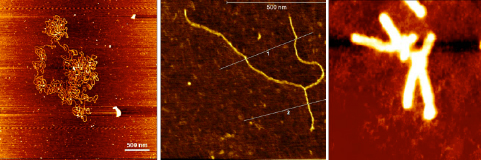
Figure 3. AFM images: large DNA bacterial artificial chromosome (Left). Replication fork isolated from vertebrate cells (Center). Translocated chromosome from cancer cells (Right).
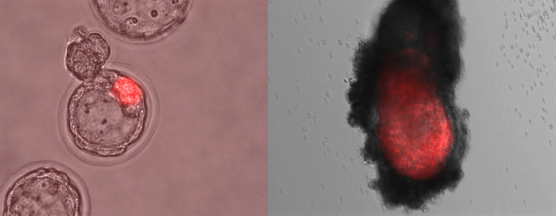
Figure 4. Phase contrast images of mouse embryos at blastocyst stage showing the localization of embryonic stem cells (marked in red) injected into morulas (left). The same embryo reimplanted into foster mothers and collected at later stage of development (right).
24 ottobre 2017 rel.02
