Functional Genomics Screening Unit

Giuseppina D’Alessandro
Born in 1988, Giuseppina D’Alessandro graduated in Medical Biotechnology at the University of Bari. After completing her studies in 2012, she moved to Milan where she obtained a PhD in Molecular Medicine at IFOM. Her doctoral research, conducted under the supervision of Dr. d’Adda di Fagagna, focused on elucidating the role of RNA and DNA:RNA hybrids in the repair of DNA double-strand breaks, one of the most deleterious type of DNA lesions. Following her PhD, Giuseppina was awarded an international fellowship through the iCARE-2 program, co-funded by AIRC and the European Union under the Marie Skłodowska-Curie Actions. This prestigious fellowship took her to the United Kingdom, where she joined the laboratory of Prof. Sir Steve Jackson at the University of Cambridge. During her postdoctoral experience at the Gurdon Institute and subsequently at the Cancer Research UK Cambridge Institute, she broadened her knowledge of DNA damage response pathways exploiting cutting-edge genome editing technologies, including CRISPR knock-out, inhibition, and base editing approaches. After five years in the UK, Giuseppina returned to Italy to establish and lead the Functional Genomics Screening Unit at IFOM. In this new role, she applies her knowledge of genome editing and CRISPR screening technologies to support IFOM scientists and to investigate DNA damage response mechanisms at scale. Throughout her career, Giuseppina has conceived and led numerous projects and coordinated collaborations with renowned international researchers. She is also a reviewer for international journals, including Nature Communications, and contributes to the preprint highlights service PreLights.
Gallery Functional genomics screening unit
Facility members
Mario Cinquanta
Marisa Aliprandi

Unit info
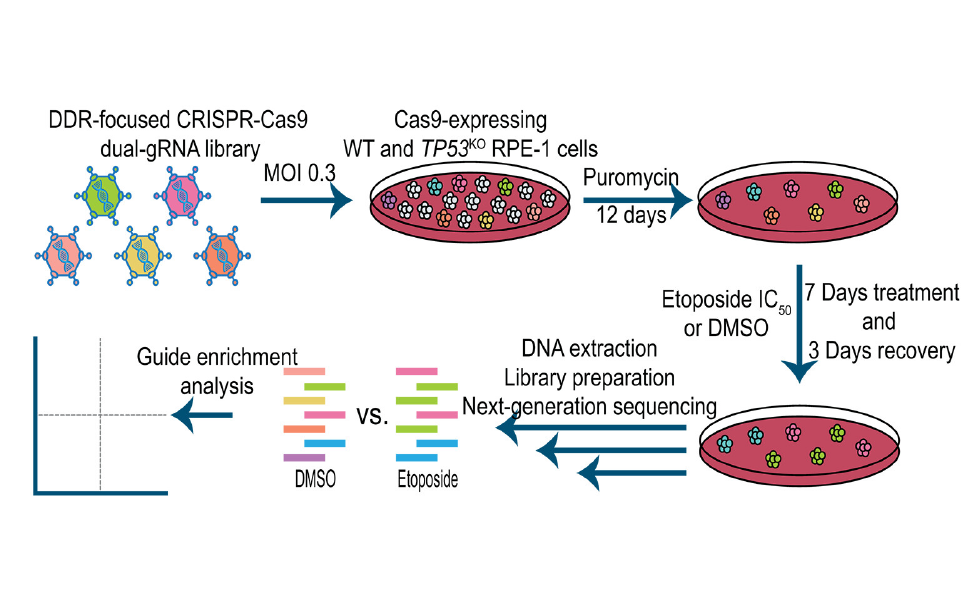
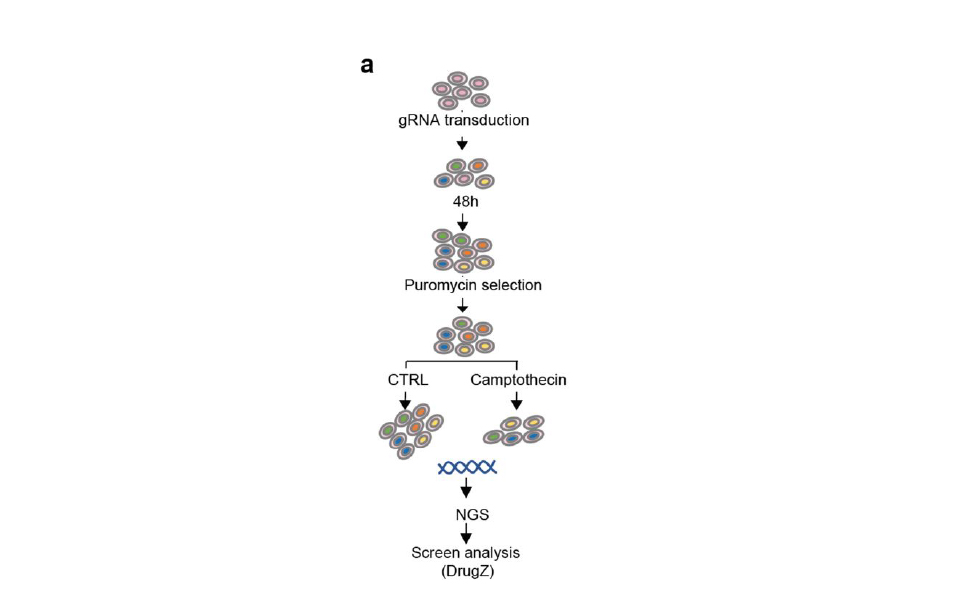
CRISPR/Cas9 is a revolutionary gene-editing tool adapted from bacterial defense mechanisms. When infected, bacteria incorporate viral DNA segments into CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) sequences. At the following viral attack, the Cas9 (CRISPR-associated protein 9) enzyme uses the RNA molecules transcribed from the CRISPR sequences to recognise the viral DNA and neutralize the threat. In 2012, Nobel Prize winners Jennifer Doudna and Emmanuelle Charpentier adapted this system for use in eukaryotic cells, creating a precise genome-editing tool that cuts DNA and relies on cellular repair mechanisms to introduce genetic changes. The Functional Genomics Screening (FGS) Unit specializes in high-throughput CRISPR screening using pooled or arrayed libraries in diverse cellular models. This technology assesses the effects of thousands of genetic perturbations at single-cell resolution with CRISPR knockout (CRISPR KO), CRISPR interference (CRISPRi), and CRISPR activation (CRISPRa). It also utilizes base and prime editing to investigate specific mutations, such as those relevant to cancer development and progression. The first approach combines a Cas module with a deaminase to convert DNA bases, while the second uses a template RNA to write the desired genetic changes into the target DNA. The FGS Unit integrates these perturbations with functional genomics readouts, including cellular viability and complex phenotypic analyses, often in collaboration with the Imaging and FACS Facility and the ETP. Comprehensive data analysis is supported by the IFOM Bioinformatics Unit. Collaborating with various IFOM groups, the FGS Unit aims to elucidate gene functions, understand their roles in disease mechanisms, and identify drug targets. In addition to supporting IFOM researchers by employing, developing and optimizing CRISPR/Cas-based tools, the Unit adopts such technologies to investigate the DNA damage response at scale.
Publications List
- Decitabine cytotoxicity is promoted by dCMP deaminase DCTD and mitigated by SUMO-dependent E3 ligase TOPORS
CJ Carnie, MJ Gotz, CS Palma-Chaundler, P Weickert, A Wanders, ...
The EMBO Journal, 1-27 (2024) - Transcription-coupled repair of DNA–protein cross-links depends on CSA and CSB
CJ Carnie, AC Acampora, AS Bader, C Erdenebat, S Zhao, E Bitensky, ...
Nature Cell Biology, 1-14 (2024) - Cockayne syndrome proteins CSA and CSB promote transcription-coupled repair of DNA-protein crosslinks independently of nucleotide excision repair
C Carnie, A Acampora, A Bader, V Gupta, G D'Alessandro, ...
Nature Research (2024) - USP37 prevents premature disassembly of stressed replisomes by TRAIP
OV Kochenova, G D'Alessandro, D Pilger, E Schmid, SL Richards, ...
bioRxiv, 2024.09.03.611025 (2024) - RAD54L2 counters TOP2-DNA adducts to promote genome stability
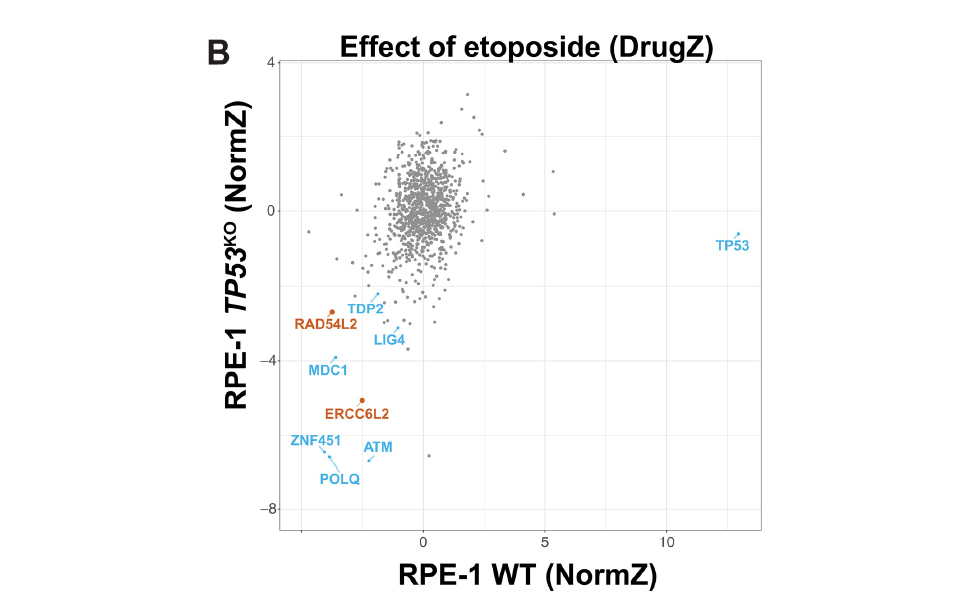
G D’Alessandro, DA Morales-Juarez, SL Richards, KC Nitiss, ...
Science Advances 9 (49), eadl2108 (2023) - The dCMP deaminase DCTD and the E3 ligase TOPORS are central mediators of decitabine cytotoxicity
CJ Carnie, MJ Gotz, CS Palma-Chaundler, P Weickert, AR Wanders, ...
bioRxiv, 2023.12.21.572728 (2023) - BRCA2 controls DNA: RNA hybrid level at DSBs by mediating RNase H2 recruitment
G D’Alessandro, DR Whelan, SM Howard, V Vitelli, X Renaudin, ...
Nature Communications 9 (1), 5376 (2018) - THE ROLE OF RNA AND DNA: RNA HYBRIDS AT DNA DOUBLE-STRAND BREAKS
G D'Alessandro
Università degli Studi di Milano (2018) - Long non-coding RNA in the control of genome stability and cancer phenotypes
G D’Alessandro, FA di Fagagna
Non-coding RNA Investigation 2 (3) (2018) - A role for RNA and DNA: RNA hybrids in the modulation of DNA repair by homologous recombination
G D’Alessandro, M Adamowicz, D Whelan, SM Howard, ...
bioRxiv, 255976 (2018) - Transcription and DNA damage: holding hands or crossing swords?
G D'Alessandro, FA di Fagagna
Journal of Molecular Biology 429 (21), 3215-3229 (2017) - A damaged genome’s transcriptional landscape through multilayered expression profiling around in situ-mapped DNA double-strand breaks
F Iannelli, A Galbiati, I Capozzo, Q Nguyen, B Magnuson, F Michelini, ...
Nature Communications 8 (1), 15656 (2017) - Rearrangements of chromosome bands 15q12-q21 are secondary to HMGA2 deregulation in conventional lipoma
G Macchia, KH Nord, G D'Alessandro, J Nilsson, L Magnusson, ...
Oncology Reports 31 (2), 807-811 (2014) - NEW FUSION GENE INVOLVING EWSR1 IN ACUTE MYELOID LEUKEMIA
G Macchia, G D'Alessandro, C Lo Cunsolo, M Carella, O Palumbo, ...
HAEMATOLOGICA 97 (s2), S133-S133 (2012)