Electron Microscopy Single Molecules
Unit info
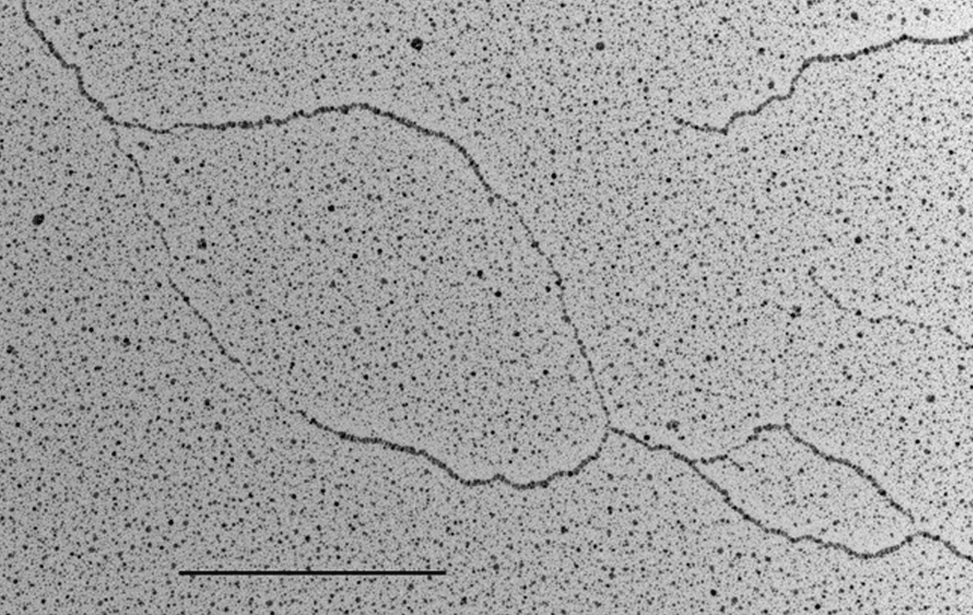
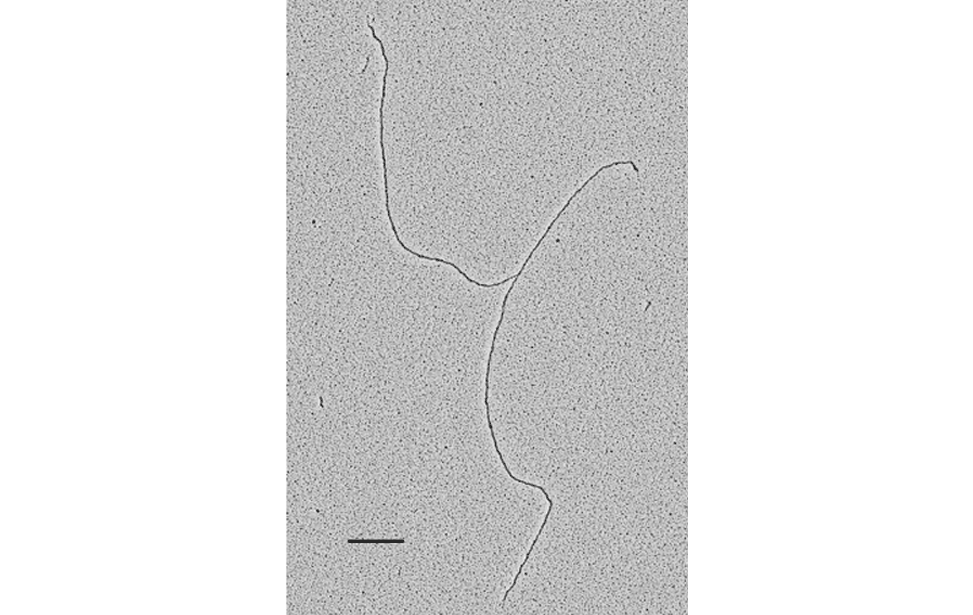
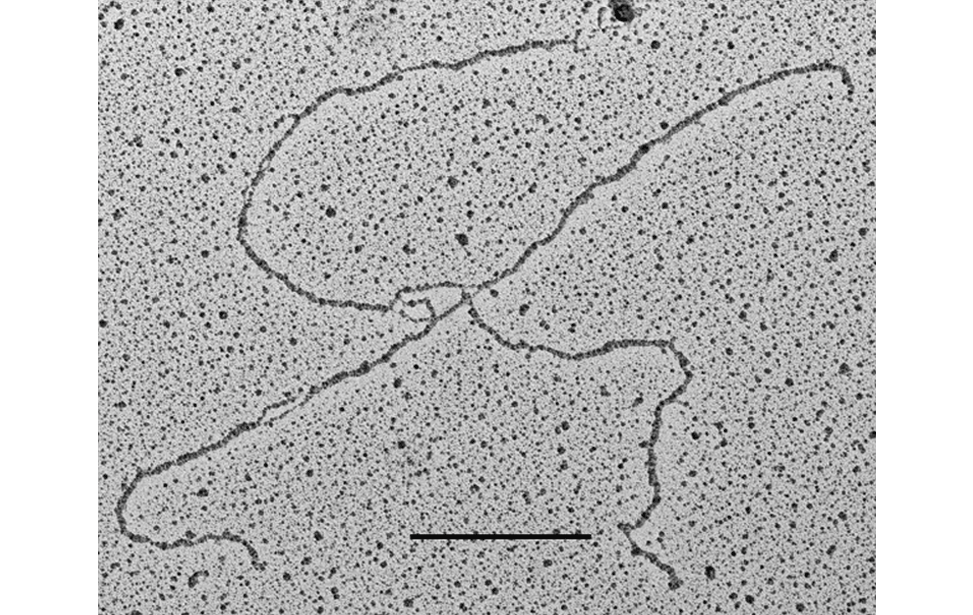
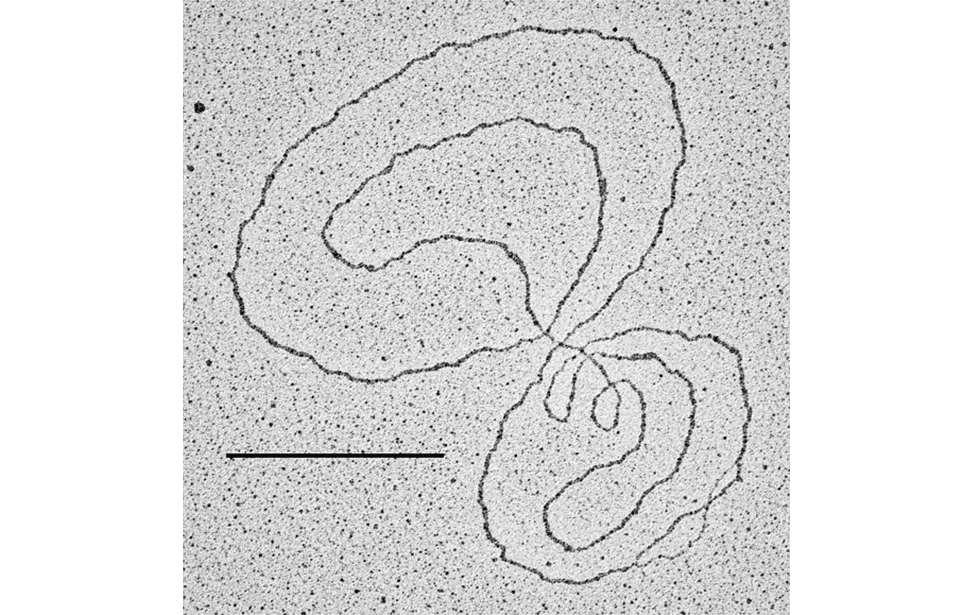
DNA metabolism reactions such as DNA replication, DNA transcription, DNA repair, DNA recombination and DNA damage tolerance are important to duplicate, express and propagate the genome in a controlled way so that cells proliferate normally and maintain genome stability. The insurgence of cancer cells is often due to failure in these DNA metabolism reactions (or of their dynamic interactions) in response to endogenous or exogenous genome threats, leading to the accumulation of cancer specific mutations in the genome. Although the fine ultra-structure of the DNA molecule is known from more than 50 years, at present, the in vivo underlaying DNA structures of the above-mentioned DNA and DNA/RNA based processes are mainly unknown. This problem can be tackled only with techniques that allow single molecule analysis with high resolution. The Technological Development Unit (TDU) uses transmission electron microscopy coupled to low angle rotary shadowing to visualize nucleic acids. This technique allows the inspection of the fine ultrastructure of in vivo chromosomal DNA intermediates on properly enriched genomic DNA samples. With one variant of this technique called “denaturing spreading” it is also possible to inspect the fine chromatin structure associated to the isolated chromosomal DNA intermediates.
The aim of the TDU is to support research projects focused on the visualization and structural analysis of in vivo chromosomal DNA and DNA-RNA intermediates involved in the duplication of the genomes. Also, the in vivo architecture of repetitive DNA sequences like centromeres, telomers, satellite DNA, the structural features of episomal DNA elements and other DNA based structures of the chromosomes can be analyzed once the corresponding genomic DNA samples have been properly enriched for those DNA sequences. Genome instability is a key feature of the cancer cells and can be associated to the accumulation of DNA, DNA-RNA and RNA structures, which are specific marks of the cancerous state. The TDU also supports the isolation and fine ultra-structural analysis of cancer associated nucleic acid structures to establish specific DNA, RNA or DNA/RNA intermediates that can eventually be used for diagnostic purposes. The TDU also provides basic techniques to stain and visualize proteins and DNA-protein complexes (through negative staining or rotary shadowing). A key activity of the TDU is the knowledge transfer towards the IFOM research groups with the aim of creating a community of experienced and independent users that actively participate to the further technological development of the TDU.

Michele Giannattasio
Michele Giannattasio was born in Milan, Italy, the 31st March 1972. He obtained the master degree in molecular biology in 1998 and the PhD in Genetics in 2002 from the University of Milan. Michele’s interest in the use of transmission electron microscopy (TEM) for the study of chromosomal DNA intermediates started in 2008 when he utilized this technique to visualize long ssDNA gaps generated during Nucleotide Excision Repair (NER). In 2010, Michele changed department at the University of Milan and became staff scientist at IFOM where he continued to apply TEM techniques to the visualization and structural analysis of chromosomal DNA intermediates in collaboration with several IFOM research groups. In these years, Michele contributed to the isolation and structural analysis of DNA damage tolerance and DNA replication intermediates and was visiting scientist at the electron microscopy research centre of the University of Zurich and the Institute of Molecular Cancer Research (Zurich), where he applied several TEM techniques and used several electron microscopes and sample preparation machines. Michele imported these TEM techniques to IFOM and made them available to IFOM scientists. In 2017, Michele became Head of the IFOM electron microscopy technological development unit (DNA/Single Molecules).
TDU organizational chart
Michele Giannattasio (Head of Electron Microscopy Unit - Single Molecules)Dana Branzei (PI of reference)
IFOM reasearch groups using/
collaborating with the TDU
Dana BranzeiVincenzo Costanzo
Fabrizio d'Adda di Fagagna
Ylli Doksani
Marco Foiani
Angela Bachi
Simona Polo
Proteins
Instruments
Application notes
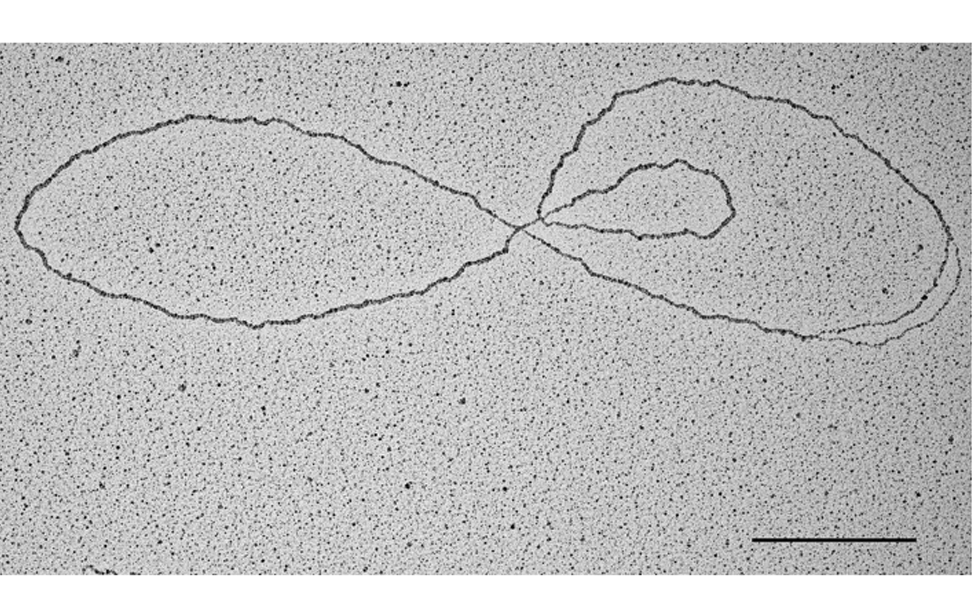
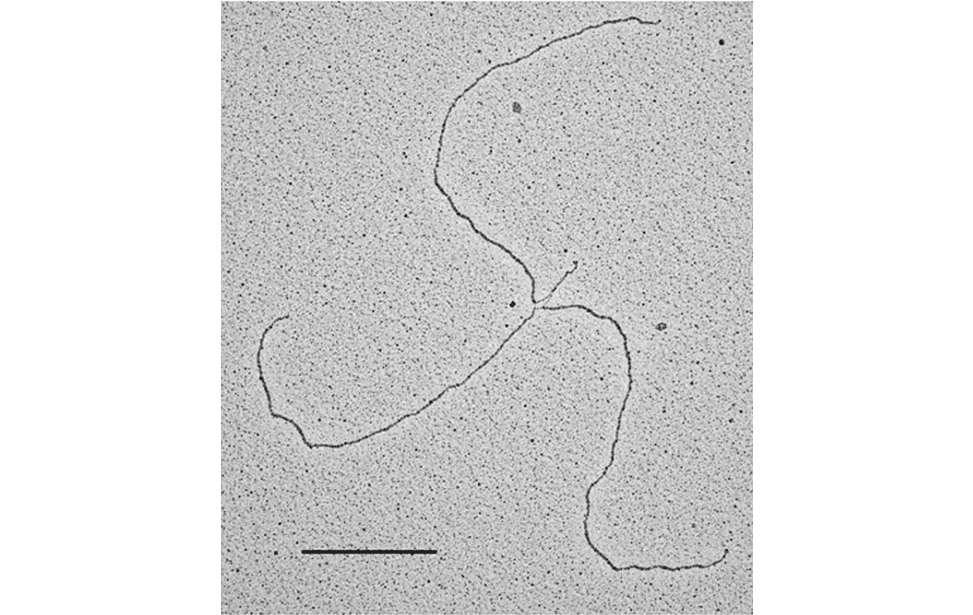
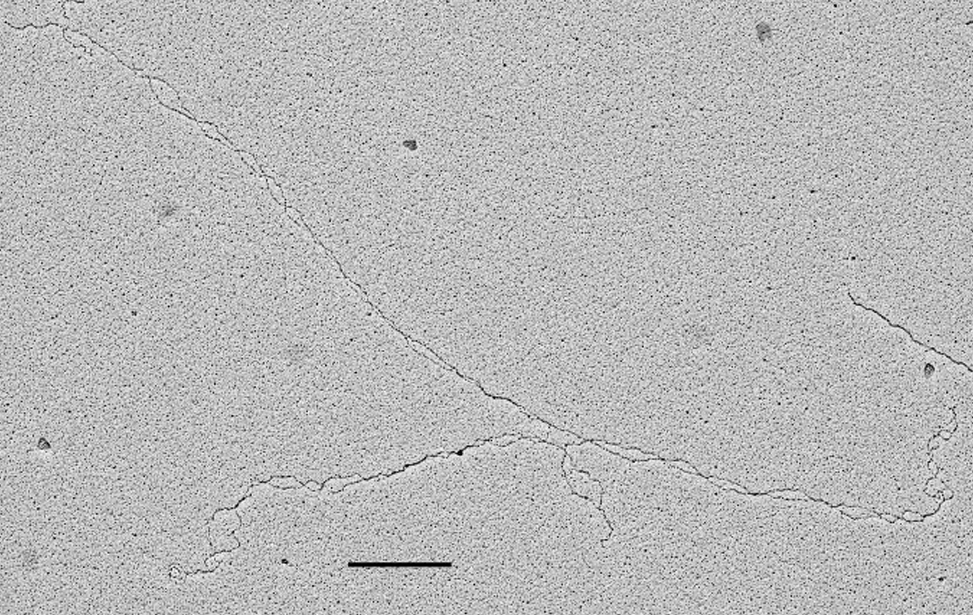
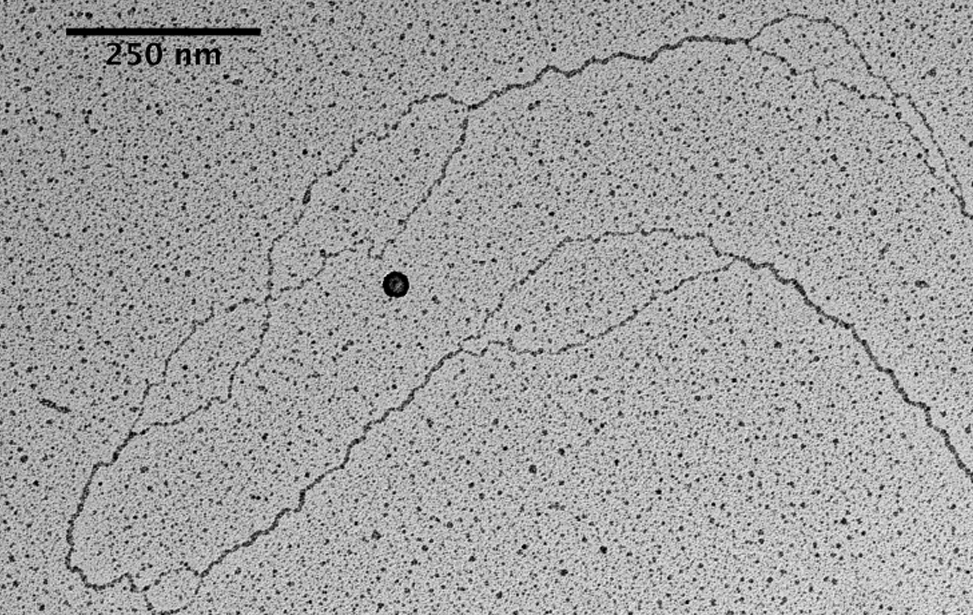
Visualization of DNA Molecules Through e-beam Evaporation and Low Angle Rotary ShadowingSelected publications including visualization data of chromosomal DNA intermediates obtained at the IFOM TDU
- Oxidative stress at telomeres triggers internal DNA loops, TRF1 dissociation, and TRF2-dependent R-loops
Nguyen TT, Mazzucco G, Kyriacou E, Lunardi T, Brandl L, Ahmed W, Doksani Y, Lingner J.
Nucleic Acids Res. 2025 Apr 10;53(7):gkaf285. doi: 10.1093/nar/gkaf285. PMID: 40219969 - HMCES corrupts replication fork stability during base excision repair in homologous recombination-deficient cells
Peña-Gómez MJ, Rodríguez-Martín Y, Del Rio Oliva M, Wijesekara Hanthi Y, Berrada S, Freire R, Masson JY, Reyes JC, Costanzo V, Rosado IV.
Sci Adv. 2025 Mar 28;11(13):eads3227. doi: 10.1126/sciadv.ads3227. Epub 2025 Mar 26.PMID: 40138423 - RAD51 protects abasic sites to prevent replication fork breakage
Hanthi YW, Ramirez-Otero MA, Appleby R, De Antoni A, Joudeh L, Sannino V, Waked S, Ardizzoia A, Barra V, Fachinetti D, Pellegrini L, Costanzo V. Mol Cell. 2024 Aug 22;84(16):3026-3043.e11. doi: 10.1016/j.molcel.2024.07.004. PMID: 39178838 Free article. - Unveiling the mechanistic link between extracellular amyloid fibrils, mechano-signaling and YAP activation in cancer.
Farris F, Elhagh A, Vigorito I, Alongi N, Pisati F, Giannattasio M, Casagrande F, Veghini L, Corbo V, Tripodo C, Di Napoli A, Matafora V, Bachi A. Cell Death Dis. 2024 Jan 11;15(1):28. doi: 10.1038/s41419-024-06424-z. PMID: 38199984 Free PMC article. - Sen1 and Rrm3 ensure permissive topological conditions for replication termination
Ramveer Choudhary, Joanna Niska-Blakie, Mohamood Adhil, Giordano Liberi, Yathish Jagadheesh Achar, Michele Giannattasio, Marco Foiani.
Cell Rep. 2023 Jul 4;42(7):112747. doi: 10.1016/j.celrep.2023.112747. - RFWD3 promotes ZRANB3 recruitment to regulate the remodeling of stalled replication forks.
Moore CE, Yalcindag SE, Czeladko H, Ravindranathan R, Wijesekara Hanthi Y, Levy JC, Sannino V, Schindler D, Ciccia A, Costanzo V, Elia AEH.
J Cell Biol. 2023 May 1;222(5):e202106022. doi: 10.1083/jcb.202106022. Epub 2023 Apr 10. PMID: 37036693 - The telomerase reverse transcriptase elongates reversed replication forks at telomeric repeats.
Huda A, Arakawa H, Mazzucco G, Galli M, Petrocelli V, Casola S, Chen L, Doksani Y.
Sci Adv. 2023 Mar 22;9(12):eadf2011. doi: 10.1126/sciadv.adf2011. Epub 2023 Mar 22. PMID: 36947627 Free PMC article. - Short-term molecular consequences of chromosome mis-segregation for genome stability
Lorenza Garribba , Giuseppina De Feudis , Valentino Martis , Martina Galli , Marie Dumont , Yonatan Eliezer , René Wardenaar , Marica Rosaria Ippolito, Divya Ramalingam Iyer, Andréa E Tijhuis , Diana C J Spierings , Michael Schubert , Silvia Taglietti , Chiara Soriani , Simon Gemble , Renata Basto , Nick Rhind , Floris Foijer , Uri Ben-David , Daniele Fachinetti , Ylli Doksani , Stefano Santaguida
Nat Commun . 2023 Mar 11;14(1):1353. doi: 10.1038/s41467-023-37095-7. - Enrichment of DNA replication intermediates by EdU pull down
Pessina, F., Romussi, A., Piccini, D., Mazzucco, G, Varasi, M., and Doksani, Y. (2022)
Methods in Cell Biology. Book Chapter - POLq prevents MRE11-NBS1-CtIP dependent fork breakage in the absence of BRCA2/RAD51 by filling lagging strand gaps.
Anjali Mann, Miguel Angel Ramirez-Otero, Anna De Antoni, Yodhara WijesekaraHanthi, Vincenzo Sannino, Giorgio Baldi, Lucia Falbo, Anna Schrempf, Sara Bernardo, Joanna Loizou, Vincenzo Costanzo.
Mol Cell 2022. Volume 82, Issue 22,2022,Pages 4218-4231.e8,ISSN 1097-2765, https://doi.org/10.1016/j.molcel.2022.09.013. - Rad51-mediated replication of damaged templates relies on monoSUMOylated DDK kinase
Chinnu Rose Joseph, Sabrina Dusi, Michele Giannattasio, Dana Branzei
Nat Commun 2022. May 5;13(1):2480. doi: 10.1038/s41467-022-30215-9. - Parental histone deposition on the replicated strands promotes error-free DNA damage tolerance and regulates drug resistance
Valeria Dolce, Sabrina Dusi, Michele Giannattasio, Chinnu Rose Joseph, Marco Fumasoni, Dana Branzei
Genes Dev 2022. Feb 1;36(3-4):167-179. doi: 10.1101/gad.349207.121. Epub 2022 Feb 3. - A rapid method to visualize human mitochondrial DNA replication through rotary shadowing and transmission electron microscopy
Martin Kosar, Daniele Piccini, Marco Foiani, Michele Giannattasio
Nucleic Acids Research, Volume 49, Issue 21, 2 December 2021, Page e121, https://doi.org/10.1093/nar/gkab770 - REV1-Polζ maintains the viability of homologous recombination-deficient cancer cells through mutagenic repair of PRIMPOL-dependent ssDNA gaps
Angelo Taglialatela, Giuseppe Leuzzi , Vincenzo Sannino , Raquel Cuella-Martin , Jen-Wei Huang , Foon Wu-Baer , Richard Baer , Vincenzo Costanzo , Alberto Ciccia.
Mol Cell. 2021 Oct 7;81(19):4008-4025.e7. doi: 10.1016/j.molcel.2021.08.016. - Checkpoint-mediated DNA polymerase ε exonuclease activity curbing counteracts resection-driven fork collapse.
Grazia Pellicanò, Mohammed Al Mamun, Dolores Jurado-Santiago , Sara Villa-Hernández , Xingyu Yin , Michele Giannattasio , Michael C Lanz , Marcus B Smolka , Joseph Yeeles , Katsuhiko Shirahige, Miguel García-Díaz , Rodrigo Bermejo
Mol Cell. 2021 Jul 1;81(13):2778-2792.e4. doi: 10.1016/j.molcel.2021.04.006. Epub 2021 Apr 30 - Smc5/6 functions with Sgs1-Top3-Rmi1 to complete chromosome replication at natural pause sites.
Sumedha Agashe , Chinnu Rose Joseph , Teresa Anne Clarisse Reyes , Demis Menolfi , Michele Giannattasio , Anja Waizenegger , Barnabas Szakal , Dana Branzei
Nat Commun. 2021 Apr 8;12(1):2111. doi: 10.1038/s41467-021-22217-w. - The human nucleoporin Tpr protects cells from RNA-mediated replication stress.
Martin Kosar , Michele Giannattasio , Daniele Piccini , Apolinar Maya-Mendoza , Francisco García-Benítez , Jirina Bartkova , Sonia I Barroso , Hélène Gaillard , Emanuele Martini, Umberto Restuccia , Miguel Angel Ramirez-Otero, Massimiliano Garre, Eleonora Verga, Miguel Andújar-Sánchez , Scott Maynard , Zdenek Hodny , Vincenzo Costanzo , Amit Kumar , Angela Bachi , Andrés Aguilera , Jiri Bartek , Marco Foiani
Nat Commun.2021 Jun 24;12(1):3937. doi: 10.1038/s41467-021-24224-3. - Telomere damage induces internal loops that generate telomeric circles.
Giulia Mazzucco, Armela Huda, Martina Galli, Daniele Piccini, Michele Giannattasio, Fabio Pessina, Ylli Doksani
Nat Commun. 2020 Oct 20;11(1):5297. doi: 10.1038/s41467-020-19139-4. - Physiological Tolerance to ssDNA Enables Strand Uncoupling during DNA Replication.
Amaia Ercilla, Jan Benada , Sampath Amitash, Gijs Zonderland, Giorgio Baldi , Kumar Somyajit, Fena Ochs ,Vincenzo Costanzo , Jiri Lukas ,Luis Toledo
Cell Rep 2020 Feb 18;30(7):2416-2429. doi: 10.1016/j.celrep.2020.01.067
- Dna2 processes behind the fork long ssDNA flaps generated by Pif1 and replication-dependent strand displacement.
Rossi SE, Foiani M, Giannattasio M.
Nature Communications. 2018 Nov 16;9(1):4830. doi: 10.1038/s41467-018-07378-5. - AND-1 fork protection function prevents fork resection and is essential for proliferation.
Abe T, Kawasumi R, Giannattasio M, Dusi S, Yoshimoto Y, Miyata K, Umemura K, Hirota K, Branzei D.
Nature Communications. 2018 Aug 6;9(1):3091. doi: 10.1038/s41467-018-05586-7. - Restoration of Replication Fork Stability in BRCA1- and BRCA2-Deficient Cells by Inactivation of SNF2-Family Fork Remodelers.
Taglialatela A, Alvarez S, Leuzzi G, Sannino V, Ranjha L, Huang JW, Madubata C, Anand R, Levy B, Rabadan R, Cejka P, Costanzo V, Ciccia A.
Molecular Cell. 2017 Oct 19;68(2):414-430.e8. doi: 10.1016/j.molcel.2017.09.036. - Fanconi-Anemia-Associated Mutations Destabilize RAD51 Filaments and Impair Replication Fork Protection.
Karina Zadorozhny, Vincenzo Sannino, Ondrej Beláň, Jarmila Mlčoušková, Mário Špírek, Vincenzo Costanzo, Lumír Krejčí.
Cell Rep 2017 Oct 10;21(2):333-340. doi: 10.1016/j.celrep. 2017.09.062. - Smarcal1-Mediated Fork Reversal Triggers Mre11-Dependent Degradation of Nascent DNA in the Absence of Brca2 and Stable Rad51 Nucleofilaments.
Kolinjivadi AM, Sannino V, De Antoni A, Zadorozhny K, Kilkenny M, Técher H, Baldi G, Shen R, Ciccia A, Pellegrini L, Krejci L, Costanzo V.
Molecular Cell. 2017 Sep 7;67(5):867-881.e7. doi: 10.1016/j.molcel.2017.07.001. Epub 2017 Jul 27. - Xenopus laevis as Model System to Study DNA Damage Response and Replication Fork Stability
Vincenzo Sannino , Federica Pezzimenti , Stefania Bertora , Vincenzo Costanzo. (2017)
Methods Enzymol. 2017;591:211-232. doi: 10.1016/bs.mie.2017.03.018. Epub 2017 Apr 8. - Centromeric DNA replication reconstitution reveals DNA loops and ATR checkpoint suppression.
Aze A, Sannino V, Soffientini P, Bachi A, Costanzo V.
Nature Cellular Biology. 2016 Jun;18(6):684-91. doi: 10.1038/ncb3344. Epub 2016 Apr 25. - Rad53-Mediated Regulation of Rrm3 and Pif1 DNA Helicases Contributes to Prevention of Aberrant Fork Transitions under Replication Stress.
Rossi SE, Ajazi A, Carotenuto W, Foiani M, Giannattasio M.
Cell Reports. 2015 Oct 6;13(1):80-92. doi: 10.1016/j.celrep.2015.08.073. Epub 2015 Sep 24. - Error-free DNA damage tolerance and sister chromatid proximity during DNA replication rely on the Polα/Primase/Ctf4 Complex.
Fumasoni M, Zwicky K, Vanoli F, Lopes M, Branzei D.
Molecular Cell. 2015 Mar 5;57(5):812-823. doi: 10.1016/j.molcel.2014.12.038. Epub 2015 Feb 5. - Visualization of recombination-mediated damage bypass by template switching.
Giannattasio M, Zwicky K, Follonier C, Foiani M, Lopes M, Branzei D.
Nature Structural & Molecular Biology. 2014 Oct;21(10):884-92. doi: 10.1038/nsmb.2888. Epub 2014 Sep 7.